confidence level: I am a physicist, not a biologist, so don’t take this the account of a domain level expert. But this is really basic stuff, and is very easy to verify.
Edit: I have added a few revisions and included a fact check of this post by an organic chemist. You can also read the comments on the EA forum to see Yudkowsky’s response.
Recently I encountered a scientific claim about biology, made by Eliezer Yudkowsky. I searched around for the source of the claim, and found that he has been repeating versions of the claim for over a decade and a half, including in “the sequences” and his TED talk. In recent years, this claim has primarily been used as an argument for why an AGI attack would be extremely deadly. I believe this claim is factually incorrect.
The quotes:
I’m going to show the various versions of the claim I found below, with the relevant sentences bolded:
To plausibly argue that “humans” were intelligently designed, you’d have to lie about the design of the human retina, the architecture of the human brain, the proteins bound together by weak van der Waals forces instead of strong covalent bonds
-Yudkowsky discussing the flaws of evolutionary design, in “the sequences” blog post “dark side epistemology”.
It was obvious years before Nanosystems that molecular nanomachines would in fact be possible and have much higher power densities than biology. I could say, “Because proteins are held together by van der Waals forces that are much weaker than covalent bonds,” to point to a reason how you could realize that after just reading Engines of Creation and before Nanosytems existed.
- Yudkowsky discussing AI interventions on the alignment forum.
A lot of the advantage of human technology is due to human technology figuring out how to use covalent bonds and metallic bonds, where biology sticks to ionic bonds and proteins held together by van der Waals forces (static cling, basically)
-Comment on a post discussing technology and AI.
Algae are tiny microns-wide solar-powered fully self-replicating factories that run on general assemblers, “ribosomes”, that can replicate most other products of biology given digital instructions. This, even though the proteins are held together by van der Waals forces rather than covalent bonds, which is why algae are far less tough than diamond (as you can also make from carbon). It should not be very hard for a superintelligence to repurpose ribosomes to build better, more strongly bonded, more energy-dense tiny things that can then have a quite easy time killing everyone.
-Yudkowsky’s example scenario for how an AI could extinct humanity, on twitter
Can you build your own synthetic biology, synthetic cyborgs? Can you blow straight past that to covalently bonded equivalents of biology where instead of proteins that fold together and are held together by static cling, you have things that go down much sharper potential energy gradients and are bundled together, people have done advanced design work about this sort of thing.
-Yudkowksy’s Ted talk, again discussing AI capabilities, during the Q&A section.
I broadly endorse this reply and have mostly shifted to trying to talk about “covalently bonded” bacteria, since using the term “diamondoid” (tightly covalently bonded CHON) causes people to panic about the lack of currently known mechanosynthesis pathways for tetrahedral carbon lattices.
-Yudkowsky’s response to my recent article a few weeks ago, talking about how to refer to potential advanced nanotechnologies.
Summarising the claim
As you can see, Yudkowsky has repeated this claim several time over a time period spanning 15 years to just a few weeks ago, in very high profile contexts.
These quotes (intentionally or unintentionally) all make roughly the same argument, which I will sum up as follows:
Proteins are held together by weak van-der-waals forces, which are weak forces, akin to static cling.
In contrast, alternatives to biological proteins could utilize strong covalent bonds, and would therefore be much more powerful.
Edit: Yudkowsky has claimed in the comments that this was not the intended message of his statements. Nonetheless, this is what he ended up saying.
Although the claim is deployed in a few different contexts, I will focus this article on the context of molecular nanotechnology, where it forms part of an argument for the potential deadliness of rogue AGI.
Bond types
Let’s start off by defining four common types of bonds. These are not the only types of bonds (for example, metallic bonds are strong but will not be discussed here). Note that the following definitions are all chem 101 simplifications. The bond strengths vary depending on which atoms are bonded and a bunch of other factors, but I just want to give a general picture here.
Covalent bonds: Covalent bonds arise when atoms “share” electron pairs between them, allowing them to get closer to a complete “shell”. These bonds are very strong, typically being hundreds of kJ/mol, depending on the atoms involved.
Ionic bonds: Some atoms really want to get rid of electrons, and some atoms really want to receive extra electrons. If one of each meets, an electron will jump from one to the other, making each atom oppositely charged, so they stick together. Ionic bond strength can vary quite a bit, with variations between 170 and 1500 kj/mol.
Hydrogen bonds: These bonds occur because hydrogen only has one electron. If that one electron covalently bonds with another atom, then it’s stuck on one side of the hydrogen, so the other side is positively charged. At the same time, another nearby atom, like oxygen, might have unbonded electrons hanging out preferentially on one side. This brings the hydrogen and other atom together electrostatically, where they bond. Compared to the previous two, this is a comparatively weak bond, typically around 10 kJ/mol.
This is the name for a collection of very weak forces acting between atoms. Primarily these are a result of fluctuating polarisations on neighbouring atoms, and tend to be transient and weak. The strength varies depending on definitions, with one source telling me 0.4 to 4 kJ/mol, and wikipedia mentioning strengths as low as 0.04 kj/mol. They are generally defined by their weakness.
A definitional note: Some sources will include hydrogen bonds as a subset of Van der Waals forces, as they both count as intermolecular forces. It is more common to separate the two definitionally, such as by saying that hydrogen bonds are permanent dipoles, while Van der Waals forces are not. The main reason to do so is that hydrogen bonds are generally much stronger and more permanent than Van der Waals forces.
Edit: Talking merely about the types of bonds can be misleading when talking about material strength, as the amount of bonds also matters. A chemist told me that “long series of hydrogen bonds or even vdW forces (in things like UHMWPE fibers) can be stronger than single covalently bound strands”. This is another problem with the quotes above.
Bonds in proteins
Now that we’ve got these definitions in place, let’s get to the question at hand. What forces are present in biology? The image below shows the primary structure of a protein chain.
What percentage of the bonds shown below correspond to each of the four bond types I listed above? I encourage you to take a guess now, before scrolling down.
The answer is as follows:
100% of the bonds are covalent.
Okay, I’ll admit, I have played a small trick here, and to explain why, I’ll have to explain a bit more about how proteins work.
To build a protein, a ribosome will read DNA to choose from a set of 22 available amino acids, and then stitch them together like beads on a string. Each amino acid is held together almost entirely by covalent bonds, and the peptide bonds between amino acids are also covalent in nature. This initial string is called the “primary structure”. The R1, R2 and R3 in the above picture represent “side chains”, extra bits that are unique to each amino acid and can have a significant effect on it’s behavior. These side chains are also primarily (or possibly entirely?) covalent in nature.
However, a long string is not a stable structure. As soon as this bead is spit out, it starts wiggling around and folding up until it’s reached a relatively stable 3d structure. The “protein folding problem” of predicting how this fold would occur was a super-hard problem, which was recently cracked using deep learning techniques by Deepminds Alphafold 2 program, which can now predict 3d structures from base structures with very high accuracy. Folding structure is often divided up into three or four regimes of structure as shown in the following image:
Okay. So we’ve established that primary structure is overwhelmingly covalent, let’s move on to the “secondary structure”. This is where the chain folds up into itself so that the “backbone” (the none side-chain part) of one section bonds with the backbone of the other. This usually takes the shape of either parallel sheets or helices, as shown in the image below.
Feel free to estimate once again, for the next image, how the bond types are divided up:
I did a rough count, and in the sheet pictured it’s roughly 90% covalent bonds. The other 10% are hydrogen bonds. It looks like a similar proportion applies in the helix, although the picture doesn’t show the other side.
Up at the tertiary level, everything becomes a lot less predictable: proteins can fold every which way, to make a huge variety of structures, which depend a lot on the individual amino acid side chains that make up the structure. The following image shows a few ways that the protein is pinned together at a higher level.
I’m not going to ask to predict the bonds this time, because I think it’s highly material dependent at this point.
We can see that in tertiary structure, side chains of the main protein can bond to each other in a ton of different ways. We can have ionic bonds, hydrogen bonds, Van der Waals interactions, and… yep, covalent bonds, in the form of disulfide links. These can act as sort of “pins”, locking folded structures in place.
This website has a nice 3d model that lets you play around and explore the different types of bonds in a complex protein.
So at long last, we have all four types of bonds available, and the proportions of each is just gonna depend on the exact sequence of amino acids used. However, for most if not all proteins, the order of prevalence will be covalent bonds>hydrogen bonds> everything else.
To summarize, proteins are held together by a combination of bond types, primarily covalent bonds and secondarily hydrogen bonds, with other forces playing some part, depending on the protein.
Edit: I think that the previous summary was a little muddled. I would summarize it now as follows:
The primary structure of protein is held together with covalent bonds, with the secondary backbone held together by hydrogen bonds. At the tertiary level, these chains are held together by a combination of many different bonds and forces, such as hydrophobic interactions, hydrogen bonds, Van der Waals forces, and covalent bond links, all of which contribute to it’s 3D structure with differing importance depending on the material.
As a result, I am now certain that the statement “proteins are held together by van der Waals forces rather than covalent bonds” is false. It’s false even if you put hydrogen bonding in the “Van der Waals forces” category, which would be misleading in this context. Nobody who knew about the actual structure of proteins would use the phrase “covalently bonded alternatives to biology”. The entire field of organic chemistry arises from carbon’s thirst for covalent bonds.
Weirdly, I’ve popped open my copy of “engines of creation”, which Yudkowsky cites for his claims, and I see nothing about Van der Waals forces or covalent bonds. In fact, Drexler spends quite a while praising the flexibility and versatility of proteins: he views custom designed protein machines as the first step on the road to nanotech. He merely notes that biological material are not as durable as metals and diamond. Of course, Drexler is not a biologist so even if he had said it, it would still have been worth fact-checking.
“Covalent bonds” were not the motivation for molecular nanotech. Rather, it was about atomically precise manufacturing: that you could avoid the wiggly randomness of biology and instead place molecules atom by atom mechanically using a system of nanoscale manipulators. The hope was that you could build a universal assembler that could build anything, and not be limited to biological materials or biological assembly techniques. This would include extremely strong structures such as diamond or pure metals, but could also be anything else that MNT proponents can dream of. If you wanted a new car, you’d just send it off to a universal assembler, which would build the entire thing from scratch in your garage.
Of course, it’s easy to dream up such things, and much harder to actually build it. As I described in a previous post, attempts to actually build such a thing have completely stalled at the first hurdle. It may just be unworkable in practicality.
Gullibility filters
I think a key danger of these sort of mistakes is that they act as a gullibility filter.
Anybody with a chemistry or biology background who hears someone confidently utter the phrase “covalently bonded equivalents to biology” will immediately have their bullshit alarm triggered, and will probably dismiss everything else you say as well. This also goes for anyone with enough skeptical instinct to google claims to ensure they have the bare minimum of scientific backing.
This isn’t an isolated incident either, by the way. See my write-up on the errors in the math in the quantum physics sequences, or the errors in economics described here.
So the people who know things, and the people who actually google things, will disproportionately come to the conclusion that EA is talking nonsense and not join, whereas the people who blindly accept any scientific sounding word they hear will just walk right in. I think this is not good for the epistemic health of a community.
Strong biology?
Moving on, what do I think of the broader points about biology being “weak”? Well it’s not like biology has slept on the idea of “make strong things”:
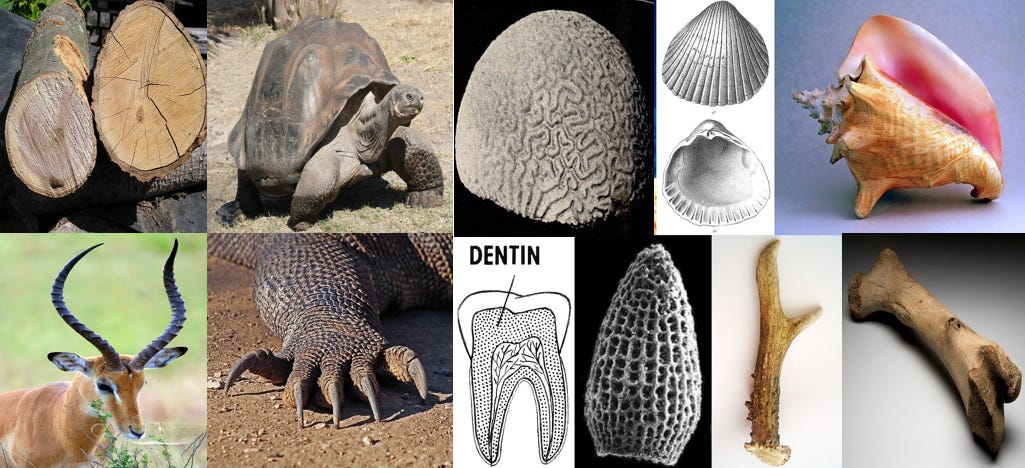
Have you heard of wood? Horns? claws, shells, bones? They all seem pretty strong to me. There are also materials with extremely high strength to weight ratios, like spider silk. Some of these are made with hard proteins such as keratin, some are made with mineralized tissue, or a combination the two.
This is not to say that a fully diamondoid based nanobot, (if such a thing is even possible) wouldn’t be stronger than these examples. Protein is not entirely covalent, which introduces weaknesses. When put into extreme conditions such as high heat, they may lose their secondary and tertiary structure and unfold back to their primary covalently bonded structure, in a process called “denaturation”. Diamond can avoid this fate as all of it’s bonds are equally strong.
My point here is merely to say that this is not a case of diamond vs “static cling”, but diamond vs wood or bone. Don’t underestimate biological ingenuity!
Flexibility
My other point is this: Are we sure we even want our proteins to be strictly covalently bonded?
In my previous post, I covered the woes of trying to stick two carbon atoms to a carbon surface. The issue was this: you have to pick the atoms up and drop them in place. But the only way to pick them up, at that scale, was to covalently bond them to your transport tip, then covalently bond the atoms and the tip to the surface, and then forcibly rip the transport atom from the surface to leave .The analogy I used was that it was like trying to lay bricks when your gloves are coated in superglue.
In this scenario, the fact that covalent bonds are strong is the problem. It would actually be fantastic if you could use weak forces here: you lightly stick the atoms to your transport tip, drop it into the covalent bond spot, and then easily pull the transport tip away.
Biology pretty much does this. For example, enzymes like the one pictured below hold molecules in place through noncovalent bonding, facilitate a chemical reaction between them, and then release them.
As part of the process, parts of the enzyme actually shift and mold their structures around the incoming molecules in order to better catalyse reactions. I’m not sure how easily you could replicate this using stiff strictly covalent structures. Edit: read the response by an organic chemist below for further discussion on this point.
Part of the reason that proteins are so flexible and versatile is exactly the fact that they aren’t fully welded together by covalent bonds, and thus can utilise multiple strengths of force as necessary for their purposes.
Back to the drawing board?
Imagine there was a boss and inventor who knew nothing about biology, and set out to build molecular machines. After fifty years of hard work, he rushes in and we get the following exchange:
Inventor: “EUREKA! I’ve invented a fully functioning self-replicating molecular machine. We can use this elaborate structure called DNA to encode strings of amino acids, and they will automatically fold into a virtually unlimited variety of 3 dimensional structures that can do a huge variety of microscale tasks, including replicating themselves using these incredibly complicated structures called “cells”. The possibilities are endless!”
Boss: “eh, I dunno, this design seems like weak shit. It’s not as tough as diamond and it’s not entirely covalent. I reckon you should abandon this design entirely and go back to the drawing board, it can’t be that hard to build something better”.
He may be right that something better is possible. But that doesn’t mean that something better can be built quickly. The DNA/RNA model has been refined with over 3 billion years of practical trial and error experimentation on a planetary scale. Evolution may be dumb and slow, but it’s also got a ridiculous head start.
I do not expect evolution to have created the best molecular machinery it is possible to ever make, given infinite time and resources. Evolution provides constraints on how changes can occur, so if you remove these constraints, you can, almost by definition, do better. But it seems to me like the smart thing to do, if you want to build effective self replicating nanomachines quickly, is to piggyback off the natural experimentation of biology, and make even cooler and better things within the DNA framework, rather than starting from scratch and trying to reinvent the wheel. I think this is true for humans, and I think it’s also true for non-godlike AI’s.
I am very excited to see what can be accomplished in biology in the years to come. We are now in the age of Alphafold 2 and CRISPR, and I expect at least some incredibly cool stuff to come out of that. In my previous post, I highlighted DNA robots and DNA origami as impressive advances that I will keep an eye on, as we learn it’s advantages and limitations .
I think there is still a lot of room to explore what biology tells us about the potential capabilities and dangers of molecular machines, biological or otherwise. I would be very interested to hear from any biologists who have expertise in these areas.
Conclusion:
I believe that the claims about protein bonding repeatedly made by Yudkowsky are factually incorrect. Proteins are primarily held together by covalent bonds, and secondarily by hydrogen bonds, with Van der Waals forces sometimes but not always contributing. Edit: Proteins are held together by a variety of forces, with a primary structure of covalent bonds, and also contain covalent bonds at the tertiary level, including sometimes as the dominant structural force. It is true that molecular machines based on diamondoid or metal could be physically stronger that biology, but attempts to build such machines have completely stalled. Biology can still build very strong materials such as wood and bone, and the use of non-covalent bonds can actually be very useful for the flexibility and versatility of proteins such as enzymes.
Edit: a breakdown of the claims I have a problem with
I originally left this as a comment, but decided to include it as an edit in the post as well. I thought that the problems with the original quotes were self-evident, but it may be worth breaking down each quote in turn. The following are ranked from roughly most to least wrong.
human technology figuring out how to use covalent bonds and metallic bonds, where biology sticks to ionic bonds and proteins held together by van der Waals forces
This is just straight up, explicitly false. Biology does not “stick to ionic bonds and proteins”. As I pointed out, biology is made up of covalent bonds at it’s very core, and uses them all the time.
covalently bonded equivalents of biology where instead of proteins that fold together and are held together by static cling, you have things that go down much sharper potential energy gradients and are bundled together
The phrase “covalently bonded equivalents to biology” implicitly states that biology is not covalently bonded. This is false.
I have mostly shifted to trying to talk about “covalently bonded” bacteria
The context of this claim is that Yudkowsky is trying to come up with a new name for deadly Drexler-style nanomachines. He has chosen “covalently bonded bacteria”, implying that “covalently bonded bacteria” and normal bacteria are different things. Except that’s not true, because bacteria is completely full of covalent bonds.
Algae are tiny microns-wide solar-powered fully self-replicating factories that run on general assemblers, “ribosomes”, that can replicate most other products of biology given digital instructions.
Okay, I just saw this one, but ribosomes are not “general” assemblers, and they cannot replicate “most other products of biology”. They do literally one thing, and that is read instructions and link together amino acids to form proteins.
For the next two, let’s establish the principle that if you say “X is held together by Y instead of Z”, you are implicitly making the statement that “X is not held together by Z”, or perhaps that “Z is irrevelant compared to Y when talking about how X is held together”, or that “Y is the dominant structural force compared to Z”. Otherwise you would not have used the word instead of. Would you utter the phrase “animal bodies are held together by flesh instead of skeletons?”
proteins bound together by weak van der Waals forces instead of strong covalent bonds
This implicitly makes the statement “proteins are not held together by strong covalent bonds”, which is false. Or it could be saying “strong covalent bonds are irrelevant compared to van der waals forces when talking about how proteins are held together”, which is also false. edit: Or it is saying that “van der waals forces are the dominant structural force in proteins”, which is also false, because this is materially dependent, and some proteins have covalent disulfide links as their dominant structural force.
even though the proteins are held together by van der Waals forces rather than covalent bonds, which is why algae are far less tough than diamond
“rather than” means the same thing as “instead of”, and therefore makes an implicitly false statement for the reason I said in the last quote.
proteins are held together by van der Waals forces that are much weaker than covalent bonds
Actually, this one is defensible, because it didn’t use the phrase “instead of”. I would still prefer more qualifying terms such “the weakest link”. If this had been the only statement, I would not have written this post.
It is fairly easy to fix most of these statements. As an example, you could say something like “the 3d structure of protein contains weak links like hydrophic bonds that are easy to break apart, whereas Drexler style tech could be made from 100% densely packed strictly covalent bonds”
I appreciate the difficulty of science communication and the need for simplification, but I believe that if it is easy to avoid saying false things, you should do so.
Final edit:
An organic chemist going by “skillissuer” reached out, saying they broadly agree with the post, but had nitpicks. I thought it was interesting enough to just include what they wrote directly here:
I think that chemistry 101 classification of bonds is a tad useless here. Instead, you can go from first principles: there are things that happen when atomic orbitals overlap (covalent bonds, metallic and such), there are interactions that are mostly electrostatic in nature (ionic, dipole-dipole, quadrupole-quadrupole—important biologically as pi-stacking, also ion-quadrupole etc) and there are things that are a result of exchange interaction (van der Waals and steric repulsion). Hydrogen bonds would be a mix of dipole-dipole and van der Waals interaction. You don’t have to transfer electrons in order to have ionic interaction, most of the time in biologically relevant situations it’s proton transfer, or charges just were there previously. Hydrophobic interactions are almost entirely a solvent effect and aren’t a bond strictly speaking
In water, i’m pretty sure that proteins are mostly held by hydrogen bonds and hydrophobic interactions. EY is correct in that some proteins hold shape by mostly noncovalent interactions, but these are mostly hydrogen, ionic, hydrophobic interactions and the proteins that actually provide mechanical strength run in continuous covalent strands through entire length of them anyway (collagen, keratin). I don’t think that counting bonds and saying that something is 90% bound covalently is a meaningful metric, because long series of hydrogen bonds or even vdW forces (in things like UHMWPE fibers) can be stronger than single covalently bound strand, ie if you tried to pull out a single strand of kevlar or collagen from bulk material, above certain length you won’t pull it apart, you’d just break it because collective energy of hydrogen bonds will be greater than single covalent bond holding it together, that’s why these fibers are strong in the first place
There is another kind of flexibility that you haven’t mentioned: proteins are made out of single covalently bound strand, yes, but these aren’t straight C-C chains. Making and especially breaking C-C bonds in controlled way is hard, proteins can be just hydrolyzed at amide bonds. If protein breaks in some way, and in real world everything breaks, it can be recycled into aminoacids (+ any cofactors etc) and then put back in a pretty straightforward way; you can’t do this with diamondoids, when it breaks, it breaks hard, and you’re done unless you’re picking everything apart atom by atom which would be much harder and more energy intensive. As it happens you can buy bulk adamantane, but it’s just made in conditions where C-C bonds are weak (high temperature) and it’s preferentially formed because it’s most stable thermodynamically among its isomers (that are starting materials). Conversely, if you use weaker bonds, you can make pieces conform to some template, or to each other without breaking everything at once—this is basis of dynamic combinatorial chemistry. There’s also entire field of self-healing materials that is based almost entirely on these either noncovalent or reversible covalent bonds
As part of the process, parts of the enzyme actually shift and mold their structures around the incoming molecules in order to better catalyse reactions. I’m not sure how easily you could replicate this using stiff strictly covalent structures.
You actually don’t have to do that, and there are some small organocatalysts that are entirely covalently bonded and do the same job. However you can’t make them from from aminoacids, these don’t have secondary structure (too small) and are generally less active. The bare minimum is to provide a receptor for transition state, and you can make it work without drastic changes in conformation. You could make your catalyst as stiff as you like, and it’ll even make activity higher—but only if none of these stiff parts interfere with binding of substrates, and your options are limited. It’s often better to leave some wiggle room. Short peptides aren’t really stiff enough in ways that matter there and instead it’s secondary and tertiary structure that puts important bits in the right place
Why is flesh weaker than diamond? Diamond is made of carbon-carbon bonds. Proteins also have some carbon-carbon bonds! So why should a diamond blade be able to cut skin?
I reply: Because the strength of the material is determined by its weakest link, not its strongest link. A structure of steel beams held together at the vertices by Scotch tape (and lacking other clever arrangements of mechanical advantage) has the strength of Scotch tape rather than the strength of steel.
Or: Even when the load-bearing forces holding large molecular systems together are locally covalent bonds, as in lignin (what makes wood strong), if you’ve got larger molecules only held together by covalent bonds at interspersed points along their edges, that’s like having 10cm-diameter steel beams held together by 1cm welds. Again, barring other clever arrangements of mechanical advantage, that structure has the strength of 1cm of steel rather than 10cm of steel.
Bone is stronger than wood; it runs on a relatively stronger structure of ionic bonds, which are no locally weaker than carbon bonds in terms of attojoules of potential energy per bond. Bone is weaker than diamond, then, because… why?
Well, partially, IIUC, because calcium atoms are heavier than carbon atoms. So even if per-bond the ionic forces are strong, some of that is lost in the price you pay for including heavier atoms whose nuclei have more protons that are able to exert the stronger electrical forces making up that stronger bond.
But mainly, bone is so much weaker than diamond (on my understanding) because the carbon bonds in diamond have a regular crystal structure that locks the carbon atoms into relative angles, and in a solid diamond this crystal structure is tesselated globally. Hydroxyapatite (the crystal part of bone) also tesselates in an energetically favorable configuration; but (I could be wrong about this) it doesn’t have the same local resistance to local deformation; and also, the actual hydroxyapatite crystal is assembled by other tissues that layer the ionic components into place, which means that a larger structure of bone is full of fault lines. Bone cleaves along the weaker fault line, not at its strongest point.
But then, why don’t diamond bones exist already? Not just for the added strength; why make the organism look for calcium and phosphorus instead of just carbon?
The search process of evolutionary biology is not the search of engineering; natural selection can only access designs via pathways of incremental mutations that are locally advantageous, not intelligently designed simultaneous changes that compensate for each other. There were, last time I checked, only three known cases where evolutionary biology invented the freely rotating wheel. Two of those known cases are ATP synthase and the bacterial flagellum, which demonstrates that freely rotating wheels are in fact incredibly useful in biology, and are conserved when biology stumbles across them after a few hundred million years of search. But there’s no use for a freely rotating wheel without a bearing and there’s no use for a bearing without a freely rotating wheel, and a simultaneous dependency like that is a huge obstacle to biology, even though it’s a hardly noticeable obstacle to intelligent engineering.
The entire human body, faced with a strong impact like being gored by a rhinocerous horn, will fail at its weakest point, not its strongest point. How much evolutionary advantage is there to stronger bone, if what fails first is torn muscle? How much advantage is there to an impact-resistant kidney, if most fights that destroy a kidney will kill you anyways? Evolution is not the sort of optimizer that says, “Okay, let’s design an entire stronger body.” (Analogously, the collection of faults that add up to “old age” is large enough that a little more age resistance in one place is not much of an advantage if other aging systems or outward accidents will soon kill you anyways.)
I don’t even think we have much of a reason to believe that it’d be physically (rather than informationally) difficult to have a set of enzymes that synthesize diamond. It could just require 3 things to go right simultaneously, and so be much much harder to stumble across than tossing more hydroxyapatite to lock into place in a bone crystal. And then even if somehow evolution hit on the right set of 3 simultaneous mutations, sometime over the history of Earth, the resulting little isolated chunk of diamond probably would not be somewhere in the phenotype that had previously constituted the weakest point in a mechanical system that frequently failed. If evolution has huge difficulty inventing wheels, why expect that it could build diamond chainmail, even assuming that diamond chainmail is physically possible and could be useful to an organism that had it?
Talking to the general public is hard. The first concept I’m trying to convey to them is that there’s an underlying physical, mechanical reason that flesh is weaker than diamond; and that this reason isn’t that things animated by vitalic spirit, elan vital, can self-heal and self-reproduce at the cost of being weaker than the cold steel making up lifeless machines, as is the price of magic imposed by the universe to maintain game balance. This is a very natural way for humans to think; and the thing I am trying to come in and do is say, “Actually, no, it’s not a mystical balance, it’s that diamond is held together by bonds that are hundreds of kJ/mol; and the mechanical strength of proteins is determined by forces a hundred times as weak as that, the part where proteins fold up like spaghetti held together by static cling.”
There is then a deeper story that’s even harder to explain, about why evolution doesn’t build freely rotating wheels or diamond chainmail; why evolutionary design doesn’t find the physically possible stronger systems. But first you need to give people a mechanical intuition for why, in a very rough intuitive sense, it is physically possible to have stuff that moves and lives and self-repairs but is strong like diamond instead of flesh, without this violating a mystical balance where the price of vitalic animation is lower material strength.
And that mechanical intuition is: Deep down is a bunch of stuff that, if you could see videos of it, would look more like tiny machines than like magic, though they would not look like familiar machines (very few freely rotating wheels). Then why aren’t these machines strong like human machines of steel are strong? Because iron atoms are stronger than carbon atoms? Actually no, diamond is made of carbon and that’s still quite strong. The reason is that these tiny systems of machinery are held together (at the weakest joints, not the strongest joints!) by static cling.
And then the deeper question: Why does evolution build that way? And the deeper answer: Because everything evolution builds is arrived at as an error, a mutation, from something else that it builds. Very tight bonds fold up along very deterministic pathways. So (in the average case, not every case) the neighborhood of functionally similar designs is densely connected along shallow energy gradients and sparsely connected along deep energy gradients. Intelligence can leap long distances through that design space using coordinated changes, but evolutionary exploration usually cannot.
And I do try to explain that too. But it is legitimately more abstract and harder to understand. So I lead with the idea that proteins are held together by static cling. This is, I think, validly the first fact you lead with if the audience does not already know it, and just has no clue why anyone could possibly possibly think that there might even be machinery that does what bacterial machinery does but better. The typical audience is not starting out with the intuition that one would naively think that of course you could put together stronger molecular machinery, given the physics of stronger bonds, and then we debate whether (as I believe) the naive intuition is actually just valid and correct; they don’t understand what the naive intuition is about, and that’s the first thing to convey.
If somebody then says, “How can you be so ignorant of chemistry? Some atoms in protein are held together by covalent bonds, not by static cling! There’s even eg sulfur bonds whereby some parts of the folded-spaghetti systems end up glued together with real glue!” then this does not validly address the original point because: the underlying point about why flesh is more easily cleaved than diamond, is about the weakest points of flesh rather than the strongest points in flesh, because that’s what determines the mechanical strength of the larger system.
I think there is an important way of looking at questions like these where, at the final end, you ask yourself, “Okay, but does my argument prove that flesh is in fact as strong as diamond? Why isn’t flesh as strong as diamond, then, if I’ve refuted the original argument for why it isn’t?” and this is the question that leads you to realize that some local strong covalent bonds don’t matter to the argument if those bonds aren’t the parts that break under load.
My main moral qualm about using the Argument From Folded Spaghetti Held Together By Static Cling as an intuition pump is that the local ionic bonds in bone are legitimately as strong per-bond as the C-C bonds in diamond, and the reason that bone is weaker than diamond is (iiuc) actually more about irregularity, fault lines, and resistance to local deformation than about kJ/mol of the underlying bonds. If somebody says “Okay, fine, you’ve validly explained why flesh is weaker than diamond, but why is bone weaker than diamond?” I have to reply “Valid, iiuc that’s legit more about irregularity and fault lines and interlaced weaker superstructure and local deformation resistance of the bonds, rather than the raw potential energy deltas of the load-bearing welds.”
Minor point about the strength of diamond:
While it is true that the ultimate strength of diamond is much higher than bone, this is relevant primarily for its ability to resist continuously applied pressure (as is its hardness enabling cutting). The point about fault lines seems more relevant for toughness, another material property that describes how much energy can be absorbed without breaking, and there bone beats diamond easily—diamond is brittle.
There are materials that have both high strength and toughness, e.g. nacre and some metallic glass, both of which are composites.
What does this operationalize as? Presumably not that if we load a bone and a diamond rod under equal pressures, the diamond rod breaks first? Is it more about if we drop sudden sharp weights onto a bone rod and a diamond rod, the diamond rod breaks first? I admit I hadn’t expected that, despite a general notion that diamond is crystal and crystals are unexpectedly fragile against particular kinds of hits, and if so that modifies my sense of what’s a valid metaphor to use.
As an physicist who is also an (unpublished) SF author, if I was trying to describe an ultimate nanoengineered physically strong material, it would be a carbon-carbon composite, using a combination of interlocking structures made out of diamond, maybe with some fluorine passivization, separated by graphene-sheet bilayers, building a complex crack-diffusing structure to achieve toughness in ways comparable to the structures of jade, nacre, or bone. It would be not quite as strong or hard as pure diamond, but a lot tougher. And in a claw-vs-armor fight, yeah, it beats anything biology can do with bone, tooth, or spider silk. But it beats it by less than an order of magnitude, far less that the strength ratio between a covalant bond to a van der Vaals bond (or even somewhat less than to a hydrogen bond). Spider silk actually gets pretty impressively close to the limit of what can be done with C-N covariant bonds, it’s a very fancy piece of evolved nanotech, with a different set of anti-crack tricks. Now, flesh, that’s pretty soft, but it’s primarily evolved for metabolic effectiveness, flexibility, and ease of growth rather than being difficult to bite through: gristle, hide, chitin, or bone spicules get used when that’s important.
But yes, if I was giving a lecture to non-technical folks where “diamond is stronger than flesh-and-bone” was a quick illustrative point rather then the subject of the lecture, I might not bother to mention that, unless someone asked “doesn’t diamond shatter easily?”, to which the short answer is “crystaline diamond yes, but nanotech can and will build carbon-carbon composites out of diamond that don’t”.
I see the appeal of using “static cling” as a metaphor to non-technical folks, but it is something of an exaggeration for hydrogen bonds—that’s significantly weaker van der Vaals bonds. “Glue” might be a fairer analogy than “static cling”. The non-protein-chain bonds in biology that are the weak links that tend to fail when flesh tears are mostly hydrogen bonds, and the quickest way to explain that to someone non-technical would be “the same sort of bonds that hold ice together”. So the proportionate analogy is probably “diamond is a lot harder than ice, and the way the human body is built, outside of a few of the strongest bits like bones, teeth and sinews, is basically held together mostly by the same sort of weakish bonds that hold ice together”.
I think it definitely means that the bone breaks first if you load them under equal pressures, and less confidently that the diamond rod breaks first if you suddenly shock it. As an example elsewhere, you don’t want to make the cores of swords out of brittle materials due to them breaking, which is why cast iron sucks. You want a ductile core that is tough (so it can absorb the energy of impacts) and a hard-but-possibly-brittle exterior (so that you can cut and keep an edge).
Plausibly this means you don’t want structural materials to be made out of diamond even if you’d want your teeth to be, at least unless you needed to sustain high loads. It looks to me like (some, stuff like femurs) bone is optimized to be flexible.
I checked this, and this post is correct. At least, when you’re talking about bones and common, natural diamonds, which are monocrystalline.
The toughness of bone is about 2-4 MPa√m(depending on the exact form of toughness) and can increase to 3-20 MPa√m locally as when bones crack, microfractures can deflect the crack from growing along the directions of maximum tensile stress.
As compared to common natural forms of diamond, which only have a toughness of 2 MPa√m. Which is mediocre compared to other engineering materials. However! Other naturally occuring forms of diamond, such as Carbando, are much tougher and just as hard. Carbando’s strength comes from the random orientation of microdiamonds i.e. it is not mono-crystaline. There’s little numerical data in the literature on this, but it is predicted that its toughness will exceed 10-20 MPa√m (paywalled article with a confusing preview). Some evidence for their toughness comes from industrial usage for thing like deep-drilling bits, unlike regular diamond. Moreover, designed dimaonds have achieved Pareto improvements in toughness and hardness compared to common natural diamonds (reaching upto 26.6MPa√mfor nanotwinned diamond).
So diamonds can be clearly superior to bone. And yeah, these things probably aren’t bound together on a large scale by van der waals forces (I haven’t looked into that aspect for unusual diamonds like Carbando, not an expert, just took a couple solid state physics courses in uni). But. Carbando seems to gain its strength from irregularities. Sometimes irregularities make materials much stronger, sometimes much weaker. Sometimes “fault lines” can be beneficial, because they allow the material to be ductile, which you want. Like the ductility of steel, IIRC, comes from irregularities in the lattice structure which are moved around as the material deforms.
And in that deformation (of a metal or other crystal), you both create the discontinuities (esp. dislocations) that increase strength while also introducing brittleness (work hardening). But the highest strength you can get with this kind of process is still not as high as you’d get from a defect free crystal, such as a monocrystalline whisker.
This is an interesting one. I’d also have thought a priori that your strategy of focusing on strength (we’re basically focusing pretty hard on tensile strength I think?) would be nice and simple and intuitive.[1]
But in practice this seems to confuse/put off quite a few people (exemplified by this post and similar). I wonder if focusing on other aspects of designed-vs-evolved nanomachines might be more effective? One core issue is the ability to aggressively adversarially exploit existing weaknesses and susceptibilities… e.g. I have had success by making gestures like ‘rapidly iterating on pandemic-potential viruses or other replicators’[2]. I don’t think there’s a real need to specifically invoke hardnesses in a ‘materials science’ sense. Like, ten pandemics a month is probably enough to get the point across, and doesn’t require stepping much past existing bio. Ten pandemics a week, coordinated with photosynthetic and plant-parasitising bio stuff if you feel like going hard. I think these sorts of concepts might be inferentially closer for a lot of people. It’s always worth emphasising (and you do), that any specific scenario is overly conjunctive and just one option among many.
If I had to guess an objection, I wonder if you might feel that’s underplaying the risk in some way?
It brings to mind the amusing molecular simulations of proteins and other existing nanomachines, where everything is amusingly wiggly. Like, it looks so squishy! Obviously things can be stronger than that.
By ‘success’ I mean ‘they have taken me seriously, apparently updated their priorities, and (I think) in a good and non-harmful way’
“Pandemics” aren’t a locally valid substitute step in my own larger argument, because an ASI needs its own manufacturing infrastructure before it makes sense for the ASI to kill the humans currently keeping its computers turned on. So things that kill a bunch of humans are not a valid substitute for being able to, eg, take over and repurpose the existing solar-powered micron-diameter self-replicating factory systems, aka algae, and those repurposed algae being able to build enough computing substrate to go on running the ASI after the humans die.
It’s possible this argument can and should be carried without talking about the level above biology, but I’m nervous that this causes people to start thinking in terms of Hollywood movie plots about defeating pandemics and hunting down the AI’s hidden cave of shoggoths, rather than hearing, “And this is a lower bound but actually in real life you just fall over dead.”
When people are highly skeptical of the nanotech angle yet insist on a concrete example, I’ve sometimes gone with a pandemic coupled with limited access to medications that temporarily stave off, but don’t cure, that pandemic as a way to force a small workforce of humans preselected to cause few problems to maintain the AI’s hardware and build it the seed of a new infrastructure base while the rest of humanity dies.
I feel like this has so far maybe been more convincing and perceived as “less sci-fi” than Drexler-style nanotech by the people I’ve tried it on (small sample size, n<10).
Generally, I suspect not basing the central example on a position on one side of yet another fierce debate in technology forecasting trumps making things sound less like a movie where the humans might win. The rate of people understanding that something sounding like a movie does not imply the humans have a realistic chance at winning in real life just because they won in the movie seems, in my experience with these conversations so far, to exceed the rate of people getting on board with scenarios that involve any hint of Drexler-style nanotech.
After reading Pope and Belrose’s work, a viewpoint of “lots of good aligned ASIs already building nanosystems and better computing infra” has solidified in my mind. And therefore, any accidentally or purposefully created misaligned AIs necessarily wouldn’t have a chance of long-term competitive existence against the existing ASIs. Yet, those misaligned AIs might still be able to destroy the world via nanosystems; as we wouldn’t yet trust the existing AIs with the herculean task of protecting our dear nature against the invasive nanospecies and all such. Byrnes voiced similar concerns in his point 1 against Pope&Belrose.
Gotcha, that might be worth taking care to nuance, in that case. e.g. the linked twitter (at least) was explicitly about killing people[1]. But I can see why you’d want to avoid responses like ‘well, as long as we keep an eye out for biohazards we’re fine then’. And I can also imagine you might want to preserve consistency of examples between contexts. (Risks being misconstrued as overly-attached to a specific scenario, though?)
Yeah… If I’m understanding what you mean, that’s why I said,
And I further think actually having a few scenarios up the sleeve is an antidote to the Hollywood/overly-specific failure mode. (Unfortunately ‘covalently bonded bacteria’ and nanomachines also make some people think in terms of Hollywood plots.) Infrastructure can be preserved in other ways, especially as a bootstrap. I think it might be worth giving some thought to other scenarios as intuition pumps.
e.g. AI manipulates humans into building quasi-self-sustaining power supplies and datacentres (or just waits for us to decide to do that ourselves), then launches kilopandemic followed by next-stage infra construction. Or, AI invests in robotics generality and proliferation (or just waits for us to decide to do that ourselves), then uses cyberattacks to appropriate actuators to eliminate humans and bootstrap self-sustenance. Or, AI exfiltrates itself and makes oodles of
horcruxesbackups, launches green goo with genetic clock for some kind of reboot after humans are gone (this one is definitely less solid). Or, AI selects and manipulates enough people willing to take a Faustian bargain as its intermediate workforce, equips them (with strategy, materials tech, weaponry, …) to wipe out everyone else, then bootstraps next-stage infra (perhaps with human assistants!) and finally picks off the remaining humans if they pose any threat.Maybe these sound entirely barmy to you, but I assume at least some things in their vicinity don’t. And some palette/menu of options might be less objectionable to interlocutors while still providing some lower bounds on expectations.
admittedly Twitter is where nuance goes to die, some heroic efforts notwithstanding
An attempt to optimize for a minimum of abstractness, picking up what was communicated here:
How could an ASI kill all humans? Setting off several engineered pandemics a month with a moderate increase of infectiousness and lethality compared to historical natural cases.
How could an ASI sustain itself without humans? Conventional robotics with a moderate increase of intelligence in planning and controlling the machinery.
People coming in contact with that argument will check its plausibility, as they will with a hypothetical nanotech narrative. If so inclined, they will come to the conclusion that we may very well be able to protect ourselves against that scenario, either by prevention or mitigation, to which a follow-up response can be a list of other scenarios at the same level of plausibility, derived from not being dependent on hypothetical scientific and technological leaps. Triggering this kind of x-risk skepticism in people seems less problematic to me than making people think the primary x-risk scenario is far fetched sci-fi and most likely doesn’t hold to scrutiny by domain experts. I don’t understand why communicating a “certain drop dead scenario” with low plausibility seems preferable over a “most likely drop dead scenario” with high plausibility, but I’m open to being convinced that this approach is better suited for the goal of x-risk of ASI being taken seriously by more people. Perhaps I’m missing a part of the grander picture?
It’s false that currently existing robotic machinery controlled by moderately smart intelligence can pick up the pieces of a world economy after it collapses. One well-directed algae cell could, but not existing robots controlled by moderate intelligence.
The question in point 2 is whether an ASI could sustain itself without humans and without new types of hardware such as Drexler style nanomachinery, which to a significant portion of people (me not included) seems to be too hypothetical to be of actual concern. I currently don’t see why the answer to that question should be a highly certain no, as you seem to suggest. Here are some thoughts:
The world economy is largely catering to human needs, such as nutrition, shelter, healthcare, personal transport, entertainment and so on. Phenomena like massive food waste and people stuck in bullshit jobs, to name just two, also indicate that it’s not close to optimal in that. An ASI would therefore not have to prevent a world economy from collapsing or pick it up afterwards, which I also don’t think is remotely possible with existing hardware. I think the majority of processes running in the only example of a world economy we have is irrelevant to the self-preservation of an ASI.
An ASI would presumably need to keep it’s initial compute substrate running long enough to transition into some autocatalytic cycle, be it on the original or a new substrate. (As a side remark, it’s also thinkable that it might go into a reduced or dormant state for a while and let less energy- and compute-demanding processes act on its behalf until conditions have improved on some metric). I do believe that conventional robotics is sufficient to keep the lights on long enough, but to be perfectly honest, that’s conditioned on a lack of knowledge about many specifics, like exact numbers of hardware turnover and energy requirements of data centers capable of running frontier models, the amount and quality of chips currently existing on the planet, the actual complexity of keeping different types of power plants running for a relevant period of time, the many detailed issues of existing power grids, etc. I weakly suspect there is some robustness built into these systems that stems not only from the flexible bodies of human operators or from practical know how that can’t be deduced from the knowledge base of an ASI that might be built.
The challenge would be rendered more complex for an ASI if it were not running on general-purpose hardware but special-purpose circuitry that’s much harder to maintain and replace. It may additionally be a more complex task if the ASI could not gain access to its own source code (or relevant parts of it), since that presumably would make a migration onto other infrastructure considerably more difficult, though I’m not fully certain that’s actually the case, given that the compiled and operational code may be sufficient for an ASI to deduce weights and other relevant aspects.
Evolution presumably started from very limited organic chemistry and discovered autocatalytic cycles based on biochemistry, catalytically active macromolecules and compartmentalized cells. That most likely implies that a single cell may be able to repopulate an entire planet that is sufficiently earth-like and give rise to intelligence again after billions of years. That fact alone certainly does not imply that thinking sand needs to build hypothetical nanomachinery to win the battle against entropy over a long period of time. Existing actuators and chips on the planet, the hypothetical absence of humans, and a HLAI or ASI moderately above it may be sufficient in my current opinion.
I rather expect that existing robotic machinery could be controlled by ASI rather than “moderately smart intelligence” into picking up the pieces of a world economy after it collapses, or that if for some weird reason it was trying to play around with static-cling spaghetti It could pick up the pieces of the economy that way too.
It seems to me as if we expect the same thing then: If humanity was largely gone (e.g. by several engineered pandemics) and as a consequence the world economy came to a halt, an ASI would probably be able to sustain itself long enough by controlling existing robotic machinery, i.e. without having to make dramatic leaps in nanotech or other technology first. What I wanted to express with “a moderate increase of intelligence” is that it won’t take an ASI at the level of GPT-142 to do that, but GPT-7 together with current projects in robotics might suffice to get the necessary planning and control of actuators come into existence.
If that assumption holds, it means an ASI might come to the conclusion that it should end the threat that humanity poses to its own existence and goals long before it is capable of building Drexler nanotech, Dyson spheres, Von Neumann probes or anything else that a large portion of people find much too hypothetical to care about at this point in time.
This totally makes sense! But “proteins are held together by van der Waals forces that are much weaker than covalent bonds” still is a bad communication.
Impressive.
“things animated by vitalic spirit, elan vital, can self-heal and self-reproduce”
Why aren’t you talking with Dr. Michael Levin and Dr. Gary Nolan to assist in the 3D mobility platform builds facilitating AGI biped interaction? Both of them would most assuredly be open to your consult forward.
Good to see your still writing.
The stuff Yudkowsky is reacting to is in ‘Nanosystems’ by Drexler. Looking in ‘Engines of Creation’ gets you the popularization, not the solid physics and chemistry. That’s all in ‘Nanosystems’, which shows how machines built out of covalently-bonded materials can be much more capable than biology. You may disagree with the arguments presented there, in which case I’d be very interested in your arguments. Unfortunately, by reacting to popularizations and tweets, you’ve inadvertently fought some straw men instead of your real opponent.
As another comment points out, when Yudkowsky says ‘proteins are held together’ he means how they are held to each other, not how they are held internally.
It’s somewhat of an exaggeration to say that proteins are held to each other by static cling. There are also hydrogen bonds. So it is more correct to say that they are held by static cling and surface tension.
This post is a stronger arguments against Drexlerian nanomachines that outperform biology in general, which doesn’t rely on the straw man.
The proteins themselves are primarily covalent, but a quick google search says that the forces in the lipid layer surrounding cells are primarily non-covalent, and the forces between cells seem also non-covalent. Aren’t those forces the ones we should be worrying about?
It seems like Eliezer is saying “the human body is a sand-castle, what if we made it a pure crystal block?”, and you’re responding with “but individual grains of sand are very strong!”
I mean… what if we did make it a pure crystal block? What would that do to the energy requirements for movement? Hydrogen bonds are pretty weak, but a cycle of “form-then-break” is pretty cheap. Covalent bonds are strong, but forming and breaking them repeatedly is energetically expensive.
Sure, but that does suggest that Yudkowsky could adjust his language a bit, right?
That doesn’t seem like the right analogy. The bonds are forced to fold over themselves because electrons repel each other and don’t want to touch. So the natural structures are mostly tetrahedral structures. Think of the vertices of a tetrahedron having edges that shoot towards and meet at the centre and you will see that these form 109° angles. When you imagine a bunch of these connected, you will see that they all start folding over themselves and will need to take up the same space which, is not possible because the electrons will repel. So you get distortions and all kinds of stuff to push them away and then it’s all complicated by a bunch of weak forces. The primary thing giving structure is this long string of covalent bonds.
Also, “forces in the lipid layer surrounding cells” are not proteins
If I understand you correctly, it seems like you have made a general argument against the existence for covalently bonded crystals. Since such structures are abundant, I don’t think much of your argument.
The structural proteins in the extracellular matrix and connective tissue (namely collagen and elastin) tend to have covalent crosslinks. So I I’m really not sure it’s accurate to say that hydrogen bonds and van der Waals forces are what’s holding the cells together.
I personally would have liked more focus on “would protein actually be better with more covalent bonds,” and less basics. But that’s because I already know the basics, and maybe the average audience doesn’t.
IMO, the more important strike against atomic-manufacturing bots is why it seems like nanobots (including cells) work better with diffusive transport and catalysis of reactions, rather than point to point transport and large energy gradients.
I think you’re modeling the audience as knowing a lot less than we do. Someone who didn’t know high school chemistry and biology would be at risk of being misled, sure. But I think that stuff should be treated as a common-knowledge background. At which point, obviously, you unpack the claim to: the weakest links in a structure determine its strength, biological structures have weak links in them which are noncovalent bonds, not all of those noncovalent bonds are weak for functional reasons, some are just hard to reinforce while constrained to things made by ribosomes. The fact that most links are not the weakest links, does not refute the claim. The fact that some weak links have a functional purpose, like enabling mobility, does not refute the claim.
Good points are made in other comments about the significance of weakest bonds, but mostly I want to say that I like this post because it’s making a clear point with clear reasoning, and was very readable.
Strong agree here, I don’t want the author to feel discouraged from posting stuff like this, it was genuinely helpful in at the very least advancing my knowledge base!
When discussing the stability of proteins, I mostly think of their folding, not whether their primary or secondary structure breaks.
The free energy difference between folded and unfolded states of a typical protein is allegedly (not an expert!) in the range 21-63 kJ/mol. So way less than a single covalent bond.
I have a friend who does his physics PhD on protein folding, and from what I remember he mostly simulates the surface charge of proteins, i.e. cares about dipole-dipole interactions (the weaker version of ionic bonds) and interaction effects with the surrounding water (again dipole-dipole afaict).
This suggests that vdW forces aren’t all that important, but the energy scale you get from imagining vdW forces is still way better than when imagining covalent bonds.
Regarding how to do enzyme-like catalysts with covalent nanotech: my first guess is that we’d want to build a structure that has several “folded”/usable states close in energy, e.g. due to rotational degrees of freedoms in the covalent bonds. This way “unfolding”/breaking the machine requires a lot of energy, while it can still mechanically move to catalyze a chemical reaction at low activation energies.
Addendum: I just learned that dipole-dipole interaction are classified as a type of vdW force in chemistry. This is different from solid state physics, where vdW is reserved for the quantum mechanical effect of induced dipole—induced dipole interaction.
So it’s indeed vdW forces that keep a protein in its shape. (This might also explain why OP found different oom for their strength?)
Another effect that is very important is determining how proteins fold is the fact that they’re dissolved in liquid water, and so hydrophilic parts of the protein want to be on the surface, while hydrophobic parts want to be on the inside, near other hydrophobic parts. This is largely an entropic force/effect.
Some other things that are true:
100% of the bonds in hydrogen gas are covalent.
Most of the fundamental particles in a water molecule are held together by the strong nuclear force, which is a much stronger binding than covalent bonds.
If you pull on a protein and stretch it apart, it looks like a long chain (maybe with a few crosslinks).
This is a non-zero amount of structure, but it looks nothing like the fully folded protein.
If we ask how the chain with the crosslinks is held together, the answer is covalent bonds that were either in the amino acids originally, or were formed when the ribosome assembled them into a chain, (or in the case of the disulfide crosslinks, were formed during the folding process).
But if we ask where the rest of the protein’s structure came from, then the answer is hydrogen bonds and hydrophobic/hydrophilic forces.
Titotal, do you agree with Eliezer’s larger point that a superintellience engineering physical actuators from the ground up can probably do much better than what our evolutionary search process produced? If so, how would you steel man the argument?
This depends a bit on what you mean with “held-together” the covalent bonds are what holds the string together but the 3D structure needs the hydrogen bonds. If you exert force on the protein the hydrogen bonds will break first and the 3D structure breaks.
The magic of protein folding is that you have a machine that can create a 2D string of amino acids gives you a 3D protein with a stable shape and reliably the same shape. For that to happen you likely inherently require that the 2D string is hold together is a way that’s strong while it wiggles around and finds the desired 3D shape. If there would be a lot of ways to create stronger bonds it would likely misfold a lot easier. And when biology does need stronger bonds Cysteine with it’s sulfide bond is available. If
Proteins are a good solution given the design constraints under which they operate, so putting them in the same category of the human retina is wrong, but I don’t think your post does a good explanation as to why.
* I studied bioinformatics but it has been a while since I have been in my molecular biology lectures.
No, strings are 1D, not 2D. Sheets are 2D.
And, of course, we can see that the ammino acid molecules that the “strings” are made of were 3D to begin with (not 2D), meaning that the strings are 3D too if you look closely enough. We can only call strings (nearly) 1D because the other two dimensions are small enough to be negligible for some purposes.
Yes, you are right.
The important aspect is that the ribosome can add one amino acid at a time to the string and once it’s finished the string can fold into the 3D shape.
I also read nanosystems and Drexlers blog. The entire concept of nanotechnology is a series of mechanical catalysts.
A mechanical catalyst is a carved out niche in the catalyst. You use nano robotics to shove in the inputs—you probably need different bonding strategies—and you press together the multiple parts of the catalyst.
Internally the shape of the catalyst allows only one possible product, and by controlling the inputs you control what gets made. (vs solution in glassware in current chemistry)
Proteins as catalysts are in fact shapes held together by many forms of bonds. This is why they are so fragile and they have failure nodes where a misfolded proteins catalyzes peers to fail. Their floppiness also allows side products and limits the complexity of the molecules that nature is able to build.
You obviously need to use a strong material for a mechanical catalyst. Diamond or platinum, etc, and its covalently bonded.
This makes the side reactions happen less often, and it theoretically could allow for higher temperature operation.
I also disagree with Yudnowsky on the feasibility of advanced AI quickly developing nanotechnology. I suspect it would require a large scale effort and a large amount of intermediate steps that a fugitive or low resource ASI would be unable to accomplish. But I think he’s correct on this claim.
I found this to be an interesting discussion, though I find it hard to understand what Yudkowsky is trying to say. It’s obvious that diamond is tougher than flesh, right? There’s no need to talk about bonds. But the ability to cut flesh is also present in biology (e.g. claws). So it’s not the case that biology was unable to solve that particular problem.
Maybe it’s true that there’s no biologically-created material that diamond cannot cut (I have no idea). But that seems to have zero relevance to humans anyway, since clearly we’re not trying to compete on the robustness of our bodies (unlike, say, turtles).
The most general possible point, that there materials that can be constructed artificially with properties not seen in biology, is obviously true, and again doesn’t seem to require the discussion of bonds.
Yeah, Yudkowsky doesn’t know what he’s talking about here.
Still, one of the ways protein engineers can make proteins more heat-stable is by adding more covalent bonds (in particular, disulfide crosslinks to prevent unfolding). See the many results for https://scholar.google.com/scholar?q=thermostability+disulfide+bond&hl=en