8 examples informing my pessimism on uploading without reverse engineering
(If you’ve already read everything I’ve written, you’ll find this post pretty redundant. See especially my old posts Building brain-inspired AGI is infinitely easier than understanding the brain, and Randal Koene on brain understanding before whole brain emulation, and Connectomics seems great from an AI x-risk perspective. But I’m writing it anyway mainly in response to this post from yesterday.)
1. Background / Context
1.1 What does uploading (a.k.a. Whole Brain Emulation (WBE)) look like with and without reverse-engineering?
There’s a view that I seem to associate with Davidad and Robin Hanson, along with a couple other people I’ve talked to privately. (But I could be misunderstanding them and don’t want to put words in their mouths.) The view says: if we want to do WBE, we do not need to reverse-engineer the brain.
For an example of what “reverse-engineering the brain” looks like, I can speak from abundant experience: I often spend all day puzzling over random questions like: Why are there oxytocin receptors in certain mouse auditory cortex neurons? Like, presumably Evolution put those receptors there for a reason—I don’t think that’s the kind of thing that appears randomly, or as an incidental side-effect of something else. (Although that’s always a hypothesis worth considering!) Well, what is that reason? I.e., what are those receptors doing to help the mouse survive, thrive, etc., and how are they doing it?
…And once I have a working hypothesis about that question, I can move on to hundreds or even thousands more “why and how” questions of that sort. I seem to find the activity of answering these questions much more straightforward and tractable (and fun!) than do most other people—you can decide for yourself whether I’m unusually good at it, or deluded.
For an example of what uploading without reverse-engineering would look like, I think it’s the idea that we can figure out the input-output relation of each neuron, and we can measure how neurons are connected to each other, and then at the end of the day we can simulate a human brain doing whatever human brains do.
Here’s Robin Hanson arguing for the non-reverse-engineering perspective in Age of Em:
The brain does not just happen to transform input signals into state changes and output signals; this transformation is the primary function of the brain, both to us and to the evolutionary processes that designed brains. The brain is designed to make this signal processing robust and efficient. Because of this, we expect the physical variables (technically, “degrees of freedom”) within the brain that encode signals and signal-relevant states, which transform these signals and states, and which transmit them elsewhere, to be overall rather physically isolated and disconnected from the other far more numerous unrelated physical degrees of freedom and processes in the brain. That is, changes in other aspects of the brain only rarely influence key brain parts that encode mental states and signals.
We have seen this disconnection in ears and eyes, and it has allowed us to create useful artificial ears and eyes, which allow the once-deaf to hear and the once-blind to see. We expect the same to apply to artificial brains more generally. In addition, it appears that most brain signals are of the form of neuron spikes, which are especially identifiable and disconnected from other physical variables.
If technical and intellectual progress continues as it has for the last few centuries, then within a millennium at the most we will understand in great detail how individual brain cells encode, transform, and transmit signals. This understanding should allow us to directly read relevant brain cell signals and states from detailed brain scans. After all, brains are made from quite ordinary atoms interacting via rather ordinary chemical reactions. Brain cells are small, and have limited complexity, especially within the cell subsystems that manage signal processing. So we should eventually be able to understand and read these subsystems.
As we also understand very well how to emulate any signal processing system that we can understand, it seems that it is a matter of when, not if, we will be able to emulate brain cell signal processing. And as the signal processing of a brain is the sum total of the signal processing of its brain cells, an ability to emulate brain cell signal processing implies an ability to emulate whole brain signal processing, although at a proportionally larger cost.
In other words:
For uploading-without-reverse-engineering (the thing I’m pessimistic about), imagine source code that looks vaguely like “Neuron 782384364 has the following intrinsic neuron property profile: {A.28, B.572, C.37, D.1, E.49,…}. It is connected to neuron 935783951 through synapse type {Z.58,Y.82,…} and to neuron 572379349 through synapse type…”.
Whereas, for uploading-with-reverse-engineering (the thing I’m more optimistic about), imagine source code that looks vaguely like an unusually complicated ML source code repository, full of human-legible learning algorithms, and other processes and stuff, but where everything has sensible variable names like “value function” and “prediction error vector” and “immune system status vector”. And then the learning algorithms are all initialized with “trained models” gathered from a scan of an actual human brain, and the parameters of those trained models are giant illegible databases of numbers, comprising this particular person’s life experience, beliefs, desires, etc.
1.2 Importantly, both sides agree on “step 1”: Let’s go get us a human connectome!
The reverse-engineering route that I prefer is:
Step 1: Measure a human connectome. The more auxiliary data, the better.
Step 2: Reverse-engineer how the human brain works.
Step 3: Now that we understand how everything works, we might recognize that our scan was missing essential data, in which case, we go back and measure it.
Step 4: Uploads!
The non-reverse-engineering route that I’m pessimistic about is:
Step 1: Measure a human connectome. The more auxiliary data, the better.
Step 2: Also do lots of measurements of neurons, organoids, etc. to fully characterize the input-output functions of neurons.
Step 3: Maybe iterate? Not sure the details.
Step 4: Uploads!
Anyway, I want to emphasize that we all agree on “step 1”. Also, I think “step 1” is the hard, slow, and expensive part, where maybe we’re building out giant warehouses full of microscopes, or whatever. So let’s do it!
If there are two disjunctive positive stories about what happens after step 1, on which people disagree, then fine! That’s all the more reason to do step 1!
(More discussion in my post Connectomics seems great from an AI x-risk perspective.)
1.3 This is annoying because I want to believe in brain uploading without reverse engineering
Let’s say we had an uninterpretable “binary blob” that could perfectly simulate a particular adult human who is very smart and nice. But there’s nothing else you can do with it. If you change random bits and try to run it, it mostly just breaks.
In terms of AGI safety, that’s a pretty great situation! We can run large numbers of sped-up people, and have them take their time to build aligned AGI, or whatever.
By contrast, let’s say we have a human upload by the reverse-engineering route. We can do the same thing, with large numbers of sped-up people. Or we can start doing extraordinarily dangerous experiments where we modify the upload to make it more powerful and enterprising. (What if we train a new one but where the cortex has 10× more neurons? And we replace all the normal innate drives with just a drive to maximize share price? Etc.)
Maybe you’re thinking: “Well, OK, but we’ll do the safe thing, not the dangerous thing.” But then I say: “What do you mean by “we”?” If there’s a top-secret project with good internal controls, then OK, sure. But if the reverse-engineering results gets published, people will do all sorts of crazy experimentation. And keeping secrets for a long time is hard. I figure that, if we can get a period where we do have uploads but don’t have non-upload brain-like-AGI, then this period would last at most a couple years, absent weird possibilities like “the uploads launch a coup”. More discussion of this is in my connectomics post.
So, I wish I believed that there was a viable path to making an uninterpretable binary blob that emulates a particular human, and can do nothing else. Alas! I don’t believe that.
2. Main text: 8 examples informing my pessimism of emulating the brain without understanding it
2.1 The mechanism by which oxytocin neurons emit synchronized pulses for milk let-down
Oxytocin neurons in the supraoptic nucleus of the hypothalamus have synchronous bursts every few minutes during suckling, which dumps a pulse of oxytocin into the bloodstream that triggers the “milk ejection reflex” (a.k.a. “milk let-down”). In his wonderful book on the hypothalamus (my review here), Gareth Leng devotes the better part of 30 pages to the efforts of his group and others to figure out how the neurons pulse:
The milk-ejection reflex had seemed to be a product of a sophisticated neuronal network that transformed a fluctuating sensory input (from the suckling of hungry young) into stereotyped bursts that were synchronized among the entire population of oxytocin cells.
…These experiments were the first convincing demonstration of a physiological role for any peptide in the brain. They did not explain the milk-ejection reflex but defined the questions that had to be answered before it could be explained. Building that explanation took another twenty years. The questions posed had no precedent in our understanding. Where did the oxytocin that was released in the supraoptic nucleus come from, if not from synapses? What triggered its release, if that release was not governed by spiking activity? What synchronized the oxytocin cells, if they were not linked by either synapses or electrical junctions? [emphasis added]
For spoilers: here’s his 2008 computational model, involving release of both oxytocin and endocannabinoids out of dendrites (contrary to the standard story where dendrites are inputs), volume transmission (as opposed to synaptic transmission), and some other stuff.
(I’m pretty sure that the word “explanation” in the quote above should be understood as “reproduction of the high-level phenomenon in a bottom-up model”, as opposed to “intuitive conceptual explanation of how that reproduction works”. I think the 2008 computational model was the first model for both of those.)
The moral of the story (I claim) is: If we’re trying to reverse-engineer the high-level behavior, it’s easy! To a first approximation, we can just say: “Well I don’t know exactly how, but these cells evidently make short pulses of oxytocin every few minutes when there are such-and-such indications of suckling”. Whereas if we’re trying to reproduce the high-level behavior (without knowing what it is) starting from properties of the individual neurons involved, then this is what seems to have taken these researchers decades of work, despite these neurons being unusually easy to access experimentally, and despite the researchers knowing exactly what they’re trying to explain.
2.2 Neurons with spike-timing-dependent plasticity, but the “timing” can involve 8-hour-long gaps
Conditioned Taste Aversion (CTA) is when you eat or drink something at time , then get nauseous at a later time , and wind up with an aversion to what you ate or drank. The interesting thing is that the aversion does not form if t₂ is just a few seconds or minutes after , nor if it’s a few days after , but it does form if it’s a few hours after .
Adaikkan & Rosenblum (2015) found a mechanism that seems to explain this. It involves neurons in the insular cortex. Novel tastes activate two molecular mechanisms (mumble mumble phosphorylation) that start 15-30 minutes after the taste, and unwind after 3 hours and 8 hours respectively. Presumably, these then interact with later nausea-related signals to enable CTA.
The moral of the story (I claim) is: If you try to characterize neurons in a controlled setting under the assumption that their behavior now depends on what was happening in the previous few seconds, but not what was happening five hours ago, then you might find that your dataset makes no sense.
As in the rest of this section, this specific example might not seem important—who cares if our uploads don’t have conditioned taste aversion?—but I suspect that this is one example of a broader category. For example, in the course of normal thinking, I think it’s easier to recall and use a concept if you were thinking about it 3 hours ago than if you haven’t thought about since yesterday. Capturing this phenomenon may be important for enabling our uploads to do good scientific research etc. I seem to recall reading a paper that suggested a molecular mechanism underlying this phenomenon (or something like it), but I can’t immediately find it.
2.3 Really weird neurons and synapses
There’s a funny thing called a “synaptic triad” in the thalamus. It’s funny because it’s three neurons connecting rather than the usual two, and it’s also funny because one of those neurons is “backwards”, with dendrites being the output rather than the input.
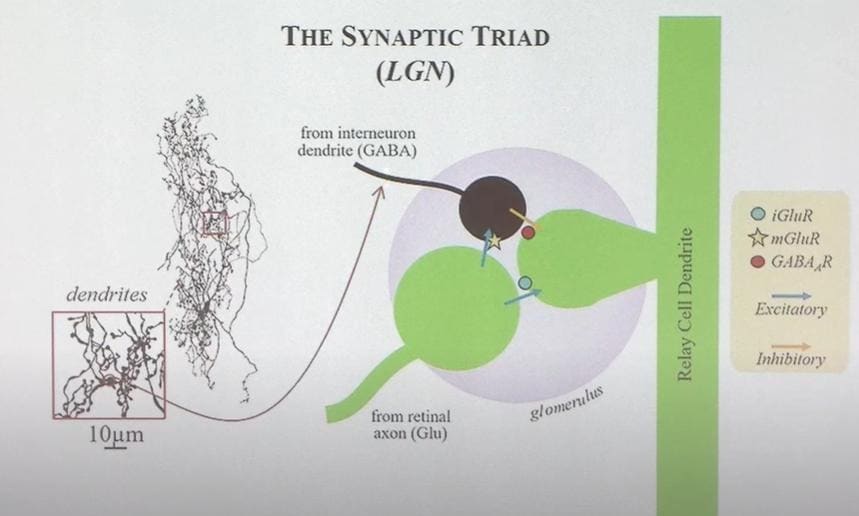
I suspect that there are many more equally weird things in the brain—this just happens to be one that I’ve come across.
The moral of the story (I claim) is: Let’s say that the uploading-without-understanding route involves separately characterizing N different things (e.g. each different type of neuron), and the reverse-engineering route involves separately characterizing M different things (e.g. each different functional component / circuit comprising how the brain’s algorithms work). Maybe you have the idea in your head that N<<M, because neurons comprise a small number of components, and they are configured into a dizzying variety of little machines throughout the brain that do different things.
If so, what I’m suggesting through this example that maybe instead N≈M, because there is also a dizzying variety of weird low-level components—i.e., N is not as small as you might think.
Separately, I think M can’t be more than maybe hundreds to low thousands, because there are only like 20,000 protein-coding genes, and they have to not only specify all the irreducible complexity of the brain’s algorithm, but also build everything else in the brain and body.
2.4 A book excerpt on simulating two particular circuits
I’m a bit hesitant to include this because I haven’t checked it, but if you trust the book The Idea of the Brain, here’s an excerpt:
…Despite having a clearly established connectome of the thirty-odd neurons involved in what is called the crustacean stomatogastric ganglion, [Eve] Marder’s group cannot yet fully explain how even some small portions of this system function. …in 1980 the neuroscientist Allen Selverston published a much-discussed think piece entitled “Are Central Pattern Generators Understandable?”...the situation has merely become more complex in the last four decades...The same neuron in different [individuals] can also show very different patterns of activity—the characteristics of each neuron can be highly plastic, as the cell changes its composition and function over time...
…Decades of work on the connectome of the few dozen neurons that form the central pattern generator in the lobster stomatogastric system, using electrophysiology, cell biology and extensive computer modelling, have still not fully revealed how its limited functions emerge.
Even the function of circuits like [frog] bug-detecting retinal cells—a simple, well-understood set of neurons with an apparently intuitive function—is not fully understood at a computational level. There are two competing models that explain what the cells are doing and how they are interconnected (one is based on a weevil, the other on a rabbit); their supporters have been thrashing it out for over half a century, and the issue is still unresolved. In 2017 the connectome of a neural substrate for detecting motion in Drosophila was reported, including information about which synapses were excitatory and which were inhibitory. Even this did not resolve the issue of which of those two models is correct.
The moral of the story (I claim) is: Building a bottom-up model that reproduces high-level behavior from low-level component neurons is awfully hard, even if we know what the high-level behavior is, and can thus iterate and iterate when our initial modeling attempts aren’t “working”. If we don’t know the high-level behavior we’re trying to explain, and thus don’t know whether our initial modeling attempts are sufficient or not—which is a necessary part of the uploading-without-understanding plan—we should expect it to be that much harder.
2.5 Failures to upload C. elegans
C. elegans only has 302 neurons, and abundant data. But we still can’t reproduce all its high-level behavior in a bottom-up model, if I understand correctly.
I’m actually pretty unfamiliar with the details, but see discussion at Whole Brain Emulation: No Progress on C. elegans After 10 Years (including the comments section).
I recall hearing somewhere [EDIT: there’s a citation here] that part of the reason that this has been a challenge is the thing I’ll talk about next:
2.6 Neurons can permanently change their behavior via storing information in the nucleus (e.g. gene expression)
See e.g. papers by Sam Gershman, David Glanzman, Randy Gallistel. Writing this section feels weird for me, because usually I’m arguing the opposite side on this topic: I subscribe to the conventional wisdom that human learning mainly involves permanent changes in and around synapses, not in the cell nucleus, and think those three people in the previous sentence go way too far in their heterodox case to the contrary. But “information storage in the nucleus is not the main story in human intelligence” (which I believe) is different from “information storage in the nucleus doesn’t happen at all, or has such a small effect that we can ignore it and still get the same high-level behavior” (which I don’t believe).
The moral of the story (I claim) is: If you don’t know what’s going on in the brain and how, then you don’t know if you’re measuring the right things, until ding, your simulation reproduces the high-level behavior. Before the agreement happens, you just have a bad model and no clue how to fix it. (Even after you’re getting agreement, you don’t know if you have sufficiently high fidelity agreement, if you don’t know what the component in question is “supposed” to do in the larger design.) I’m not sure if it’s even possible to measure gene expression etc. in human brain slices. I doubt it’s easy! But if we understand the role of gene expression in how such-and-such part of the brain works, we can reason about what if anything we’re losing by leaving it out, and see if there’s any easier workaround. If we don’t understand what it’s doing, then we’re sitting there staring at a simulation that doesn’t match the data, and we don’t know how critical a problem that is, and whether we’re missing anything else.
2.7 Glial cell gene expression that changes over hours
I was just reading about this yesterday:
Astrocytes in the [suprachiasmatic nucleus of the hypothalamus], for instance, show rhythmic expression of clock genes and influence circadian locomotor behavior (25) [source]
This is kinda a combination of Section 2.3 above (there are lots of idiosyncratic low-level components to characterize, as opposed to lots of circuits made from the same basic neuronal building blocks) and Section 2.6 above (you need to be simulating gene expression if you want to get the right high-level behavior).
2.8 Metabotropic (as opposed to ionotropic) receptors can have almost arbitrary effects over arbitrary timescales
If I understand correctly, the brain uses lots of ionotropic receptors, whose effects are to immediately and locally change the flow of ions into a neuron. Great! That’s easy to model.
Unfortunately, the brain also uses lots of metabotropic receptors (a.k.a. G-protein-coupled receptors), whose effects are—if I understand correctly—extraordinarily variable, indeed almost arbitrary. Basically, when they attach to a ligand, they then set off a signaling cascade, which can ultimately have pretty much any effect on the cell over any timescale. It might change the neuron’s synaptic plasticity rules, it might change gene expression, it might increase or decrease the cell’s production of neuropeptides, you name it. (If it’s not already obvious, these 8 examples are not mutually exclusive—many of the previous subsections involve metabotropic receptors.)
The moral of the story (I claim) is: If your goal is to get the right high-level behavior using a bottom-up model of low-level components, then you presumably need to figure out what all these signaling cascades are, experimentally, for every neuron with metabotropic receptors. Again I’m not an expert, but that seems very hard.
- Connectomics seems great from an AI x-risk perspective by (30 Apr 2023 14:38 UTC; 107 points)
- Connectomics seems great from an AI x-risk perspective by (EA Forum; 30 Apr 2023 14:38 UTC; 10 points)
- 's comment on Does davidad’s uploading moonshot work? by (4 Nov 2023 11:34 UTC; 10 points)
- 's comment on The Big Nonprofits Post by (30 Nov 2024 12:05 UTC; 4 points)
- 's comment on Straightforward Steps to Marginally Improve Odds of Whole Brain Emulation by (25 Mar 2025 1:14 UTC; 4 points)
- 's comment on Counting AGIs by (27 Nov 2024 16:11 UTC; 4 points)
- 's comment on Computational functionalism probably can’t explain phenomenal consciousness by (13 Dec 2024 15:52 UTC; 4 points)
- 's comment on What are the actual arguments in favor of computationalism as a theory of identity? by (19 Jul 2024 3:27 UTC; 1 point)
I’ve been meaning to write up something about uploading from the WBE 2 workshop, but still haven’t gotten around to it yet …
I was in a group called “Lo-Fi Uploading” with Gwern, Miron, et al, and our approach was premised on the (contrarian?) idea that figuring out all the low-level detailed neuroscience to build a true bottom-up model (as some—davidad? - seem to imagine) just seems enormously too complex/costly/far etc, for the reasons you mention (but also just see davidad’s enormous cost estimates).
So instead why not just skip all of that: if all we need is functional equivalence, aim directly for just that. If the goal is just to create safe AGI through neuroscience inspiration, then that inspiration is only useful to the extent it aids that goal, and no more.
If you take some computational system (which could be an agent or human brains) and collect a large amount of highly informative input/output data from said system, the input/output data indirectly encodes the original computational structure which produced it, and training an ANN on that dataset can recover (some subset) of that structure. This is distillation[1].
When we collect huge vision datasets pairing images/videos to text descriptions and then train ANNs on those, we are in fact partly distilling the human vision system(s) which generated those text descriptions. The neurotransmitter receptor protein distributions are many levels removed from relevance for functional distillation.
So the focus should be on creating a highly detailed sim env for say a mouse—the mouse matrix. This needs to use a fully differentiable rendering/physics pipeline. You have a real mouse lab environment with cameras/sensors everywhere—on the mice, the walls, etc, and you use this data to learn/train a perfect digital twin. The digital twin model has an ANN to control the mouse digital body; an ANN with a sufficiently flexible architectural prior and appropriate size/capacity. Training this “digital mouse” foundation model correctly—when successful—will result in a functional upload of the mouse. That becomes your gold standard.
Then you can train a second foundation model to predict the ANN params from scan data (a hypernetwork), and finally merge these together for a multimodal inference given any/all data available. No neuroscience required, strictly speaking—but it certainly does help, as it informs the architectural prior which is key to success at reasonable training budgets.
Distilling the knowledge in a neural network
On the narrow topic of “mouse matrix”: Fun fact, if you didn’t already know, Justin Wood at Indiana University has been doing stuff in that vicinity (with chicks not mice, for technical reasons):
(I haven’t read any of his papers, just listened to him on this podcast episode, from which I copied the above quote.)
2.5 sounds like we’re just way off and shouldn’t expect to get WBE until superhuman neurology AI. But given that, the remaining difficulties seem just another constant factor. The question becomes, do you expect “If you change random bits and try to run it, it mostly just breaks.” to hold up?
For Section 2.5 (C. elegans), @davidad is simultaneously hopeful about a human upload moonshot (cf. the post from yesterday) and intimately familiar with C. elegans uploading stuff (having been personally involved). And he’s a pretty reasonable guy IMO. So the inference “c. elegans stuff therefore human uploads are way off” is evidently less of a slam dunk inference than you seem to think it is. (As I mentioned in the post, I don’t know the details, and I hope I didn’t give a misleading impression there.)
I’m confused by your last sentence; how does that connect to the rest of your comment? (What I personally actually expect is that, if there are uploads at all, it would be via the reverse-engineering route, where we would not have to “change random bits”.)
Oh, okay.
My second sentence meant “If neurology AI can do WBE, a slightly (on a grand scale) more superhuman AI could do it without reverse engineering.”. But actually we could just have the AI reverse-engineer the brain, then obfuscate the upload, then delete the AI.
Suppose the company gets bought and they try to improve the upload’s performance without understanding it. My third sentence meant, would they find that the Algernon argument applies to uploads?
My suspicion is that the answer is likely no, and this is actually a partial crux on why I’m less doomy than others on AI risk, especially from misalignment.
My general expectation is that most of the difficulty is hardware + ethics, and in particular the hardware for running a human brain just does not exist right now, primarily because of the memory bottleneck/Von Neumann bottleneck that exists for GPUs, and it would at the current state of affairs require deleting a lot of memory from a human brain.
I disagree about the hardware difficulty of uploading-with-reverse-engineering—the short version of one aspect of my perspective is here, the longer version with some flaws is here, the fixed version of the latter exists as a half-complete draft that maybe I’ll finish sooner or later. :)
I separate possible tech advances by the criterion: “Is this easier or harder than AGI?” If it’s easier than AGI, there’s a chance it will be invented before AGI, if not, AGI will invent it, thus it’s pointless to worry over any thought on it our within-6-standard-deviations-of-100IQ brains can conceive of now. WBE seems like something we should just leave to ASI once we achieve it, rather than worrying over every minutia of its feasibility.
Oops, sorry for leaving out some essential context. Both myself, and everyone I was implicitly addressing this post to, are concerned about the alignment problem, e.g. AGI killing everyone. If not for the alignment problem, then yeah, I agree, there’s almost no reason to work on any scientific or engineering problem except building ASI as soon as possible. But if you are worried about the alignment problem, then it makes sense to brainstorm solutions, and one possible family of solutions involves trying to make WBE happen before making AGI. There are a couple obvious follow-up questions, like “is that realistic?” and “how would that even help with the alignment problem anyway?”. And then this blog post is one part of that larger conversation. For a bit more, see Section 1.3 of my connectomics post. Hope that helps :)
I totally agree with that notion, I however believe the current levers of progress massively incentivize and motivate AGI development over WBE. Currently regulations are based on flops, which will restrict progress towards WBE long before it restricts anything with AGI-like capabilities. If we had a perfectly aligned international system of oversight that assured WBE were possible and maximized in apparent value to those with the means to both develop it and push the levers, steering away from any risky AGI analogue before it is possible, then yes, but that seems very unlikely to me.
Also I worry. Humans are not aligned. Humans having WBE at our fingertips could mean infinite tortured simulations of the digital brains before they bear any more bountiful fruit for humans on Earth. It seems ominous, fully replicated human consciousness so exact a bit here or there off could destroy it.
I’m not sure what you’re talking about. Maybe you meant to say: “there are ideas for possible future AI regulations that have been under discussion recently, and these ideas involve flop-based thresholds”? If so, yeah that’s kinda true, albeit oversimplified.
I think that’s very true in the “WBE without reverse engineering” route, but it’s at least not obvious in the “WBE with reverse engineering” route that I think we should be mainly talking about (as argued in OP). For the latter, we would have legible learning algorithms that we understand, and we would re-implement them in the most compute-efficient way we can on our GPUs/CPUs. And it’s at least plausible that the result would be close to the best learning algorithm there is. More discussion in Section 2.1 of this post. Certainly there would be room to squeeze some more intelligence into the same FLOP/s—e.g. tweaking motivations, saving compute by dropping the sense of smell, various other architectural tweaks, etc. But it’s at least plausible IMO that this adds up to <1 OOM. (Of course, non-WBE AGIs could still be radically superhuman, but it would be by using radically superhuman FLOP (e.g. model size, training time, speed, etc.))
Hmm. I should mention that I don’t expect that LLMs will scale to AGI. That might be a difference between our perspectives. Anyway, you’re welcome to believe that “WBE before non-WBE-AGI” is hopeless even if we put moonshot-level effort into accelerating WBE. That’s not a crazy thing to believe. I wouldn’t go as far as “hopeless”, but I’m pretty pessimistic too. That’s why, when I go around advocating for work on human connectomics to help AGI x-risk, I prefer to emphasize a non-WBE-related path to AI x-risk reduction that seems (to me) likelier to actualize.
I grant that a sadistic human could do that, and that’s bad, although it’s pretty low on my list of “likely causes of s-risk”. (Presumably Ems, like humans, would be more economically productive when they’re feeling pretty good, in a flow state, etc., and presumably most Ems would be doing economically productive things most of the time for various reasons.)
Anyway, you can say: “To avoid that type of problem, let’s never ever create sentient digital minds”, but that doesn’t strike me as a realistic thing to aim for. In particular, in my (controversial) opinion, that basically amounts to “let’s never ever create AGI” (the way I define “AGI”, e.g. AI that can do groundbreaking new scientific research, invent new gadgets, etc.) If “never ever create AGI” is your aim, then I don’t want to discourage you. Hmm, or maybe I do, I haven’t thought about it really, because in my opinion you’d be so unlikely to succeed that it’s a moot point. Forever is a long time.
I agree with a lot of what you said but I am generally skeptical of any emulation that does not work from a bottom up simulation of neurons. We really don’t know about how and what causes consciousness and I think that it can’t be ruled out that something with the same input and outputs at a high level misses out on something important that generate consciousness. I don’t necessarily believe in p zombies, but if they are possible then it seems they would be built by creating something that copies the high level behavior but not the low level functions. Also on a practical level, I don’t know how you could verify a high level recreation is accurate. If my brain is scanned into a supercomputer that can run molecular dynamics on every molecule in my brain then I think there is very little doubt that it is an accurate reflection of my brain. It might not be me in the sense that I don’t have any continuity of consciousness, but it is me in the sense that it would behave like me in every circumstance. Conversely, a high level copy could miss some subtle behavior that is not accounted for in the abstraction used to form the model of the brain. If an LLM was trained on everything I ever said, it could imitate me for a good long while but it wouldn’t think the same way I do. A more complex model would be better but not certain to be perfect. How could we ever be sure that our understanding was such that we didn’t miss something subtle that emerges from the fundamentals? Maybe I’m misinterpreting the post, but I don’t see a huge benefit from reverse engineering the brain in terms of simulating it. Are you suggesting something other than an accurate simulation of each neuron and its connections? If you are, I think that method is liable to miss something important. If you are not I think that understanding each piece but not the whole is sufficient to emulate a brain.